Let us look back even further, back millions of years, to examine latest genetic studies of how Capsicum diverged from Solanaceae.
From the 2017 gochu dna study I had summarized previously, we have already examined the following chart showing the evolutionary divergence of Capsicum from Solanaceae with calculated times of divergence in millions of years.
From the above chart, we can see the evolution of diverging species took places across hundreds of millions of years, partially taking place even during the reign of dinosaurs that ended 65 million years ago.
With that much passage of time across even shifts in the Earth’s tectonic plates, the following chart shows the tremendously wide array of cultivated species that diverged from Solanaceae.
http://www.seehint.com/catalog/2014/2014_11/%EA%B0%80%EC%A7%93%EA%B3%BC.jpg
What is most unique about Capsicum is the development of the unique capsaicin compound that makes chili peppers “hot” flavor sensation. With capsaicin compound being a strong irritant to mammals, stimulating a burning sensation to mouth, eyes, and skin, it seems the compound was selective to specifically promote propagation by birds. Birds being unaffected by capsaicin, wild “bird peppers” are small with upright pods with generous seeds within to entice birds to feast and release flat hard shelled seeds across long distance flight paths.
Evolution of a hot genome
https://www.pnas.org/content/111/14/5069
- Fig. 1: The genome of chile pepper in an evolutionary context. (A) A phylogeny of lamiid angiosperms, the family Solanaceae in yellow and chile pepper in red. (B) Syntenic dotplot of tomato chromosome 2 (vertical) versus chiltepin pepper chromosomes 2 (Upper) and 4 (Lower), with syntenic gene duplicates colored according to their Ks values (synonymous substitutions per synonymous site), as plotted in the x axis of C against numbers of gene pairs on the y axis. Ks, increasing along the x axis, is a proxy for time since speciation or gene duplication. Purple, orthologous synteny (youngest); blue, paralogous synteny descending from the triplication observed in Solanaceae; green, paralogous synteny dating to the paleohexaploidy event at the base of core eudicots (oldest). Note corresponding purple, blue, and green peaks in the Ks plot, the latter two subtle.
We can see the Capsicum line eventually leading to Capsicum annuum diverged 19.6 million years ago. Capsaicin compound emerged to exploit unaffected birds as seed dispersal agents quite well before the Last Glacial Maximum that started 1.8 million years ago when North America finally connected by land bridge to South America, cutting off the ocean currents between the Atlantic and Pacific Oceans. With birds being considered descendants of the dinosaurs, nearly 20 million years of spreading capsicum pepper seeds is quite a long time before anatomically modern humans finally cultivated chili peppers.
Chromosomal evolution of the plant family Solanaceae
https://www.researchgate.net/publication/42254578_Wu_F_Tanksley_SD_Chromosomal_evolution_in_the_plant_family_Solanaceae_BMC_Genom_11_182/fulltext/0f58e4f53829b1e540929576/Wu-F-Tanksley-SD-Chromosomal-evolution-in-the-plant-family-Solanaceae-BMC-Genom-11-182.pdf?origin=publication_detail
Much of the recent investigations into sequencing a variety of capsicum peppers is a quest to find disease resistance. The CM334 wild pepper is too small to cultivate for commercial purposes but is of tremendous interest for having the highest level of disease resistance reflecting millions of years of evolution.
New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication
Genome Biology
01 November 2017
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1341-9
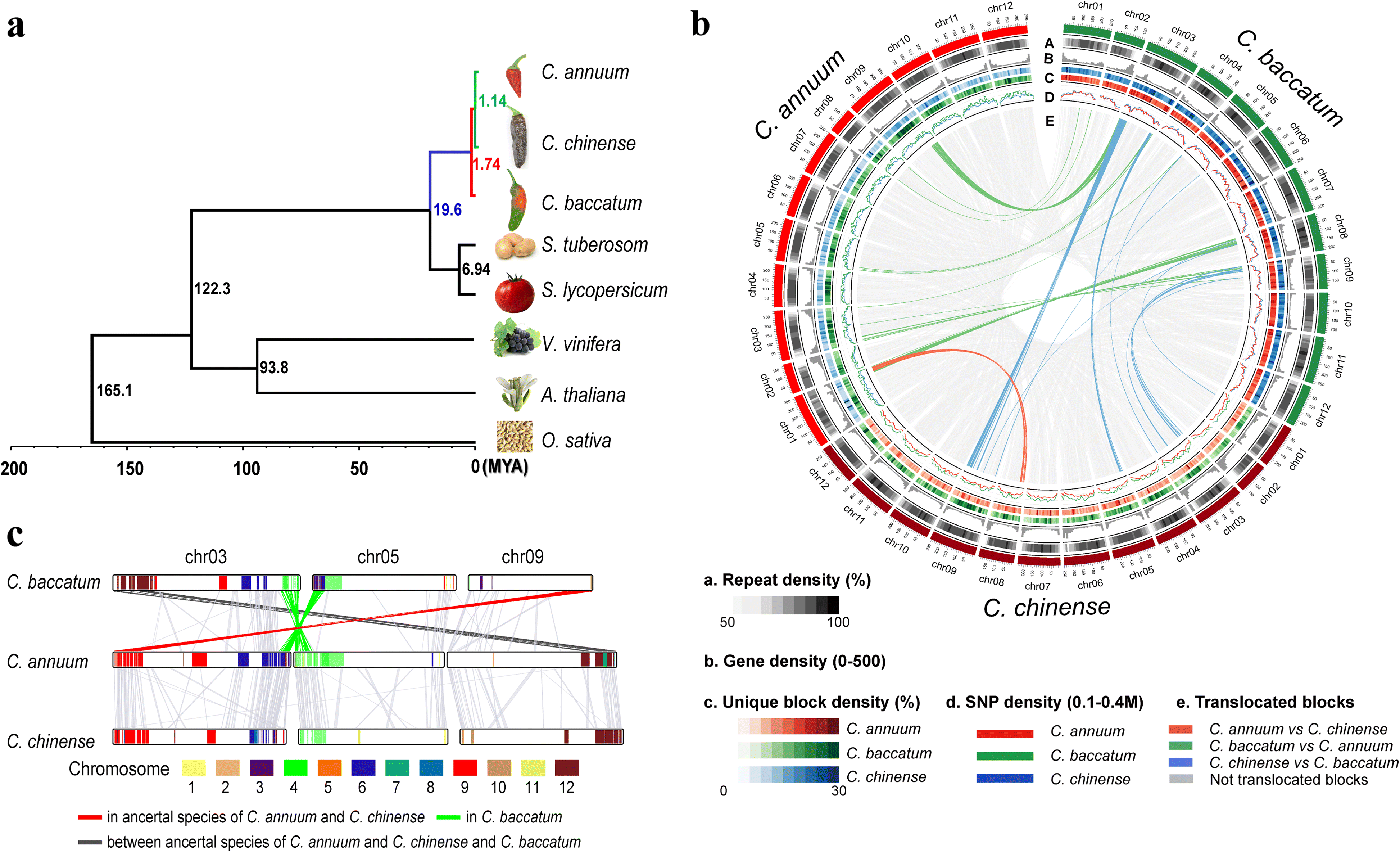
- • Figure 1: Lineage-divergence and genome structure comparisons of three Capsicum species. a The reconstructed phylogenetic tree of eight plant genomes indicates their evolutionary relationships and estimated divergence times. b The circular diagram shows the distribution of repeats, genes, genomic variations, and genome rearrangements in the pepper genomes. The subcategories indicate the density of repeats (A), genes (B), species-specific blocks (C), and SNPs (D) in the pepper genomes. The subcategory (E) depicts collinear and translocated blocks among the pepper genomes. c A linear comparison of the rearranged blocks in the pepper genomes. Colors in the bars indicate translocated regions when comparing to tomato and potato genomes. The line colors indicate translocations in the ancestral lineage leading to Annuum and Chinense (red), in Baccatum (green), and in the ancestor of Annuum and Chinense or Baccatum (dark gray)